Trojans investigate mouse and human kidneys to hasten new remedies for renal disease
A free online kidney atlas built by USC researchers empowers stem cell scientists everywhere to generate more humanlike tiny kidneys for testing new drugs and creating renal replacement therapies.
“Stem-cell based technologies hold great promise for developing kidney replacement and regeneration therapies,” said Nils Lindstrom, first author of three new studies and a research associate in Stem Cell Biology and Regenerative Medicine at the Keck School of Medicine of USC. “Getting there requires detailed knowledge of how kidneys normally form so the process can be replicated in cell cultures in the lab. Our data will help us and other scientists improve current techniques to make better tiny functional kidneys.”
Over the past four years, USC Stem Cell researchers and USC Viterbi School of Engineering computer scientists have documented the molecular, cellular and genetic similarities and differences between human and mouse kidney formation so they can find treatments for kidney disease, which affects about 30 million Americans or 15 percent of U.S. adults.
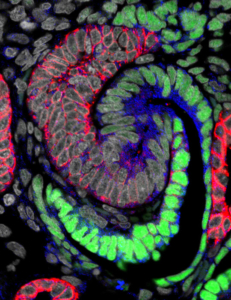
At an early stage, a nephron forming in the human kidney generates an S-shaped structure. Green cells will generate the kidneys’ filtering device, and blue and red cells specialized regions responsible for distinct nephron activities. (Photo/
Stacy Moroz and Tracy Tran,Andy McMahon Lab, USC Stem Cell)
The three-study series, published on Feb. 15 in the Journal of the American Society of Nephrology, provides the first cellular and molecular characterization of how the human kidney develops in a mother’s womb, said Andrew McMahon, study senior author and W.M. Keck Provost Professor of Stem Cell Biology and Regenerative Medicine and Biological Sciences at the Keck School of Medicine.
Kidney research has been a focus of the McMahon Lab for the past quarter century.
“Our research bridges a critical gap between animal models and human applications,” McMahon said. “The data we collected and analyzed creates a knowledge-base that will accelerate stem cell-based technologies to produce mini-kidneys that accurately represent human kidneys for biomedical screening and replacement therapies.”
The new, open-source data, available at www.gudmap.org, provides the first systematic, high-resolution atlas or databank for human kidney genesis.
How human and mouse kidneys are similar
The kidney plays a central role in controlling the body’s ecosystem by regulating blood pressure and removing waste products.
The smallest functional unit that helps remove blood waste from the body are nephrons. Nephrons are formed in the human kidney only during fetal life, before the stem cells that generate them are exhausted.
USC Stem Cell researchers analyzed the developmental differences in scale, timing and basic structure between human and mouse kidneys. They gained insight into the regulatory processes that maintain, expand and turn kidney stem cells into mature, functional kidney structures by examining human kidneys at different stages of development and juxtaposing their observations with mouse kidneys.
Researchers compared 26 human kidney “anchor genes” with their mouse equivalents. Kidney anchor genes are required for organ development. Only three genes (11 percent) have comparable expression between mouse and human kidneys: SLC22A6, ENTPD5 and UMOD.
“If the goal is to treat human kidney disease, clearly, it’s better to focus on genes that are also active in human kidneys,” McMahon said.
Intractable problems require multidisciplinary collaboration
The median wait time for a kidney transplant is 3.6 years, meaning people have to put their lives on standby for years as they go to three dialysis appointments per week.
Kidney disease is a big, intractable problem, so it requires a multidisciplinary approach. That’s why McMahon teamed up with Carl Kesselman, study co-author and dean’s professor of industrial and systems engineering at USC Viterbi.
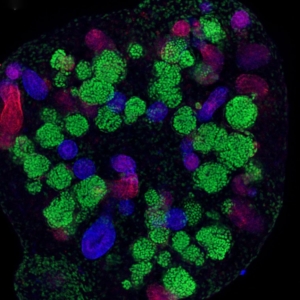
A kidney organoid on Day 16 of differentiation. The staining depicts the segmentation of nephron-like structures in the organoid at an early developmental stage. (Image/Tracy Tran, Andrew McMahon Lab, USC Stem Cell)
Kesselman and his team at the Information Sciences Institute at USC Viterbi built software that automated many of the tasks researchers needed to perform, such as recording data observed by a high-resolution microscope. The tool not only fast-tracked one of the three studies but also created an online, searchable library to help other stem cell scientists in their kidney disease research.
“If you think of data as the modern version of a book, we gave the researcher tools to write the book, made the library where the book is stored and created a catalog system so others can find the book and check it out,” said Kesselman, a principal investigator at the USC Michelson Center for Convergent Bioscience.
To achieve this, Kesselman’s team created DERIVA (Discovery Environment for Relational Information and Versioned Assets), a software program that collected data from microscopes and organized them and other study data into virtual photo albums that easily can be shared with other researchers.
“It’s the difference between taking a photo with film or taking it with your smartphone and using the metadata that is automatically generated to organize your photos,” Kesselman said. “At some point, you have so many photos that they become unmanageable. You can’t find the one you want. DERIVA automatically catalogs your data so you can analyze massive amounts of data with Zen-like calm and share them with all of your friends throughout the world.”
Jill McMahon, Jinjin Guo, Tracy Tran, Qiuyu Guo, Elisabeth Rutledge, Riana Parvez, Gohar Saribekyan, Christopher Liao, Albert Kim, Ahmed Abdelhalim, Seth Ruffins and Andrew Ransick from the Department of Stem Cell Biology and Regenerative Medicine at the Keck School of Medicine; Robert Schuler from the Information Sciences Institute at USC Viterbi; Matthew Thornton, Brendan Grubbs, Guilherme De Sena Brandine and Andrew D. Smith from USC; and Laurence Basking from the University of California, San Francisco also contributed to this study.
The research was supported by the California Institute for Regenerative Medicine (CIRM LA1-06536), National Institutes of Health (NIH DK107350, DK094526, DK110792, 5F32DK109616-02), National Institute of Diabetes and Digestive and Kidney Diseases (F31DK107216, 1U24DK110814) and Joint Medical/Research Council/Wellcome Trust (099175/Z/12/Z) as well as by a USC Stem Cell Hearst Fellowship and a USC Research Enhancement Fellowship.