A USC-led team of researchers releases expanded data set of brain scans from stroke patients with more than four times the data in the hopes of speeding up large-scale stroke recovery research.
By Sidney Taiko Sheehan
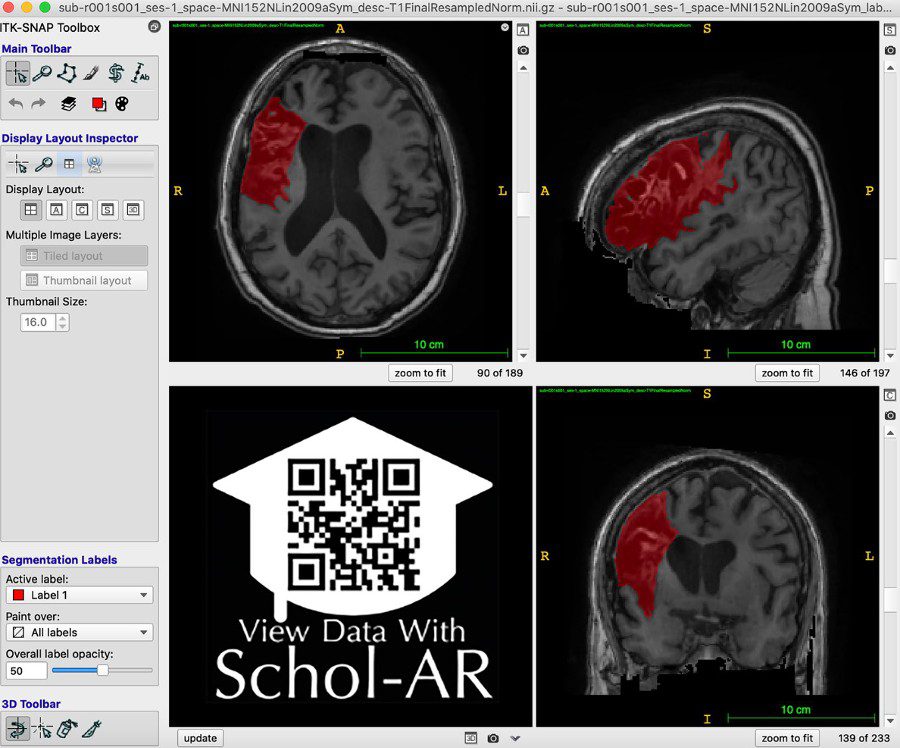
Example of lesion segmentation (red). A video example of lesion segmentation can be viewed through the Schol-AR App by scanning the QR code in the bottom left with a mobile device. Photo credit: USC Stevens INI
A major expansion of an open-source stroke neuroimaging data set known as ATLAS could give a major boost to stroke recovery research.
According to the Centers for Disease Control and Prevention, every 40 seconds someone in the U.S. has a stroke, making stroke a leading cause of disability. During a stroke, blood flow to part of the brain is cut off. Without oxygen, brain cells cease to function, causing damage to an area of the brain, known as a lesion. Lesions are detected by magnetic resonance imaging (MRI), and they are a critical aspect that researchers study as they develop, test, and implement stroke recovery programs.
Before studying a lesion, neuroanatomy experts manually draw boundaries on imaging around the lesion in a process called segmentation—a time-consuming practice that can be subjective and requires neuroanatomical expertise. Accurate lesion segmentation is critical in stroke rehabilitation research. While manual segmentation is the current gold standard, researchers hope to develop algorithms to automate the practice so they can spend more time examining lesion images with the goal of understanding how lesions impact recovery after a stroke.
Now, a newly expanded data set of brain scans from stroke patients called the Anatomical Tracings of Lesion After Stroke (ATLAS) is set to expedite the automation of lesion segmentation. Led by researchers at the Keck School of Medicine of USC’s Mark and Mary Stevens Neuroimaging and Informatics Institute (Stevens INI), the new release of ATLAS v 2.0 now includes 1,271 stroke MRI images with manually segmented lesions, according to a new paper in Scientific Data, a Nature journal. The hope is the data set will facilitate large-scale stroke recovery research which will eventually allow a clinician to predict a patient’s response to therapies based on analysis of the patient’s unique scans and data.
“We are thrilled to share this open science resource with the scientific community,” says Sook-Lei Liew, PhD, lead author of the study and associate professor with joint appointments at the Stevens INI, the Chan Division of Occupational Science and Occupational Therapy, the Division of Biokinesiology and Physical Therapy, and the USC Viterbi School of Engineering. “We hope that our public data set, along with our stroke lesion segmentation challenges, like our ISLES 2022 challenge at MICCAI, our Grand Challenge, and our educational challenge in collaboration with the RAMP studio and our colleagues in Paris will not only spur the creation of robust, powerful algorithms to move the field of stroke neuroimaging forward, but also provide useful educational opportunities for students around the world.”
The various challenges hosted by the ATLAS team invite the scientific community to participate in the process of developing and testing algorithms that attempt to automate the lesion segmentation with the greatest accuracy. The ATLAS team also showcases their collaborative strength with researchers at USC and beyond. HoSung Kim, PhD, assistant professor of neurology at the Stevens INI, created a neuroimaging analysis pipeline to help standardize the images in the data set. Tyler Ard, PhD, assistant professor of research at the Stevens INI and creator of Schol-AR, developed custom software for advanced visualization of the lesioned data set, rendering it into several extremely high-resolution videos and images. Sixteen other co-authors across the university assist with analysis, clinical characterization, and the collection and storage of data.
“Dr. Liew’s team exemplifies how researchers here at the Stevens INI approach complex medical questions from a multimodal approach,” says Stevens INI Director Arthur W. Toga, PhD. “Our faculty members team up with experts across the university and beyond to apply their expertise in machine learning, data visualization, informatics, and neuroradiology to deliver a valuable set of open-source MR images. By gathering all these images and sharing them, we are creating the optimal environment for discovery.”
Data from the project are stored by the International Neuroimaging Data-Sharing Initiative (INDI), housed at the Child Mind Institute, and by the Inter-University Consortium for Political and Social Research (ICPSR), housed at the University of Michigan. So far, thousands of researchers worldwide have downloaded the ATLAS data set.
“Algorithm development using this larger data set, with hidden test and evaluation data sets for machine learning challenges, should lead to more robust solutions. As predictive algorithms improve, our long-term goal is for clinicians to use MRI to inform decisions about stroke patients’ treatment. With the progression of this work, we hope to see a future where clinicians can analyze an individual’s data to discover their likelihood of responding to different treatments. Using a precision medicine approach, their stroke rehabilitation therapy could be personalized to maximize their recovery,” notes Liew.
For more information about accessing the ATLAS data set, visit this site.
This research is funded in part by the National Institutes of Health (R01NS115845; R25HD105583; K01HD091283; P2CHD06570; R21HD067906; R01NS090677; R00HD091375; R01NS076348-01, P20 GM109040; R01HD094731; R01HD065438). For additional funders, see publication in Scientific Data.
