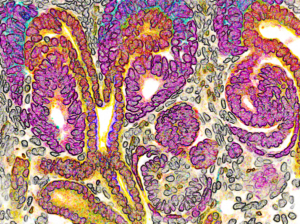
Developing human nephron, the filtering unit of the kidney (Image by Nils O. Lindstrom and Tracy Tran/McMahon Lab)
Running early or running late can have big consequences—especially when it comes to the progenitor cells involved in human kidney development. According to a new study in Developmental Cell from the USC Stem Cell laboratory of Andy McMahon, the progenitor cells that form the kidney’s filtering units, called nephrons, mature into entirely different types of cells based on when they arrive to the scene of nephron formation.
During development, about 1 million nephrons form in a human kidney. First authors Nils O. Lindstrom, Guilherme De Sena Brandine and Tracy Tran observed that every time one of these structures forms in a human kidney, the nephron progenitor cells (NPCs) gradually commit to becoming various mature cell types and joining the developing nephron. Those NPCs that arrive early into the forming nephron start to differentiate and become the “tubule,” which is the portion that controls the reabsorption of important compounds back into the blood and carries the urine to the collecting ducts. The NPCs that are recruited late into the forming nephron instead develop into the “glomerulus,” the structure that filters the blood.
“Timing is critical in determining the type of mature cell that each progenitor will become,” said Lindstrom.
Developing human nephron, the filtering unit of the kidney (Image by Nils O. Lindstrom and Tracy Tran/McMahon Lab)
To show that their predictions were accurate, Lindstrom and colleagues used genetically labelled NPCs in mouse kidneys and grew these under a microscope while capturing images with time-lapse imaging. This allowed them to demonstrate how NPCs gradually move into the newly forming nephron and turn on genes that are specific to particular cell types.
Thanks to a cross-university collaboration with Andrew D. Smith’s group in the Molecular and Computational Biology section of the Department of Biological Sciences at the USC Dornsife College of Letters, Arts and Sciences, the scientists developed new techniques to analyze and interpret what is known as “single cell RNA sequencing data.” Single cell RNA sequencing is a novel technique that allows scientists to break apart whole tissues and organs and observe gene activity in every cell. Using this approach, the team documented how NPCs turn into intermediate cell types with specific gene activity, which identifies them as the precursors to particular mature cell types.
In order to achieve the full diversity of cell types in the nephron, when attempting to grow kidneys in laboratory settings, scientists need to understand how these precursor cell types form under normal circumstances in the body. Single cell RNA sequencing provides a view of all the genes and genetic pathways that are activated when specific cell types form in the nephron.
“By studying normal human nephron development, we’re gaining important information about how to recapitulate this intricate process in the laboratory,” said McMahon, director of the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research at USC. “The hope is that laboratory-grown nephrons can be used to further study the process of development, screen potential therapies to treat disease, and eventually provide the building blocks to assemble functional kidneys for transplantation into patients.”
Additional co-authors include Andrew Ransick, Gio Suh, Jinjin Guo, Albert Dale Kim, Riana K. Parvez, Seth Walter Ruffins, Elisabeth A. Rutledge, Matthew E. Thornton, Brendan Grubbs and Jill A. McMahon from USC.
The work was supported by the National Institutes of Health (DK107350, DK094526, DK110792) and the California Institute for Regenerative Medicine (LA1-06536).
By Cristy Lytal